Mathematical tools gallery
Filter by:
In collaboration with Centre Hospitalier Universitaire de Lyon, France and University of Patras, Greece.
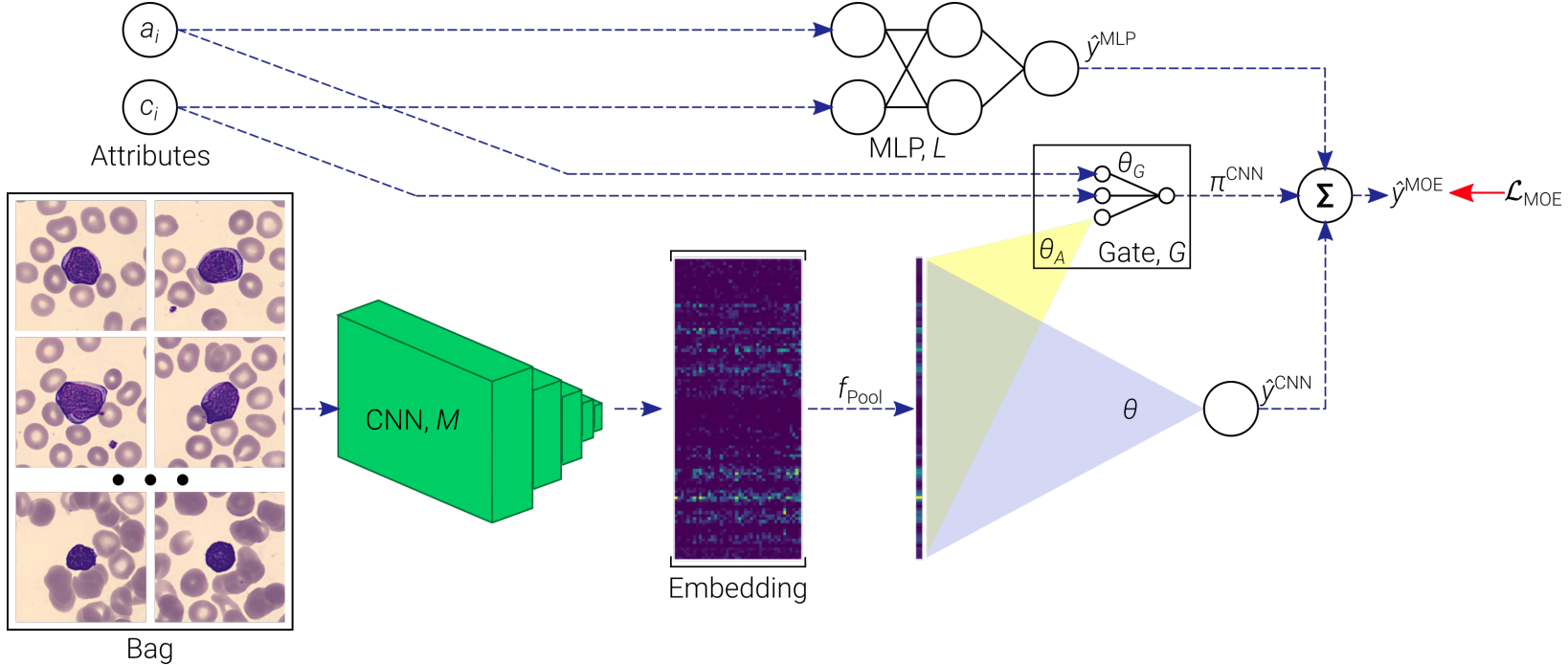
Lymphocytosis (i.e., absolute lymphocyte count above 4 × 109/L) is a common finding in patients, which can be either a reaction to infection, acute stress, and so on (termed reactive), or the manifestation of a lymphoproliferative disorder—a type of cancer of the lymphocytes (termed tumoral). In existing clinical practice, diagnosis (as either reactive or tumoral) relies on visual microscopic examination of blood smears together with the integration of clinical attributes such as age and lymphocyte count. Taking into consideration the visual assessment based on clinical attributes together with texture and size of the lymphocytes in the blood smear, a diagnosis of the subtype of lymphoid malignancy is performed. In this work, we investigate the use of recent advances in deep learning and propose an end-to-end trainable multi-instance convolutional neural network within a mixture-of-experts formulation that combines information from two types of data—blood smears and clinical attributes—for the diagnosis of lymphocytosis. The convolutional network learns to extract meaningful features from images of blood cells using an embedding level approach and aggregates them. Moreover, the mixture-of-experts model combines information from these images as well as clinical attributes to form an end-to-end trainable pipeline for diagnosis of lymphocytosis. Our results demonstrate that even the convolutional network by itself is able to discover meaningful associations between the images and the diagnosis, indicating the presence of important unexploited information in the images. The mixture-of-experts formulation is shown to be more robust while maintaining performance via. a repeatability study to assess the effect of variability in data acquisition on the predictions. Our code and datasets can be found at at this link.

The population models we consider are systems of differential equations, where each line represents the time evolution of an individual in some phase space. The statistical inference of such a model consists of determining the parameters driving the dynamics from a subset of partially observed individual trajectories. This task is challenging when the population is large and is composed of heterogeneous individuals in interaction. Such a situation frequently occurs when studying an ecosystem. We consider in particular the case of symmetric population models, i.e., when the dynamics remain invariant by permuting the labels of the individuals. This property provides a series of efficient surrogates to the dynamics, based on a distinction between macroscopic and microscopic levels during the simulation. In the eventuality of the existence of a mean-field limit, the statistical inference can be significantly simplified for large populations, taking advantage of the approximated independence of the individuals.

E-Recruitment has been widely adopted and practiced worldwide over the last ten years, which has opened up more opportunities for employers and job seekers. With the increasing amount of online recruitment data, seeking intelligent ways for automatically and effectively matching the right candidates to the right jobs is a crucial task for employers. Moreover, providing better interpretations about the matching results enhances results’ reliability and usability, facilitating related downstream tasks, such as job-oriented skill measuring, skills recommender, and next job recommendation.
In collaboration with Assistance Publique - Hopitaux de Paris hospitals network.
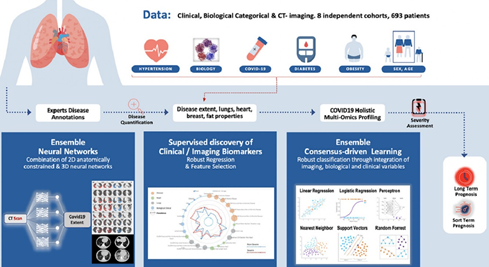
Computed tomography (CT) imaging has been proven to be an important tool for screening, disease quantification and staging. The latter is of extreme importance for organizational anticipation as well as to accelerate drug development through rapid, reproducible and quantified assessment of treatment response. In this study a multi-centric cohort of 536 was considered. We designed an AI driven scheme for the quantification of CT scans for patients suffering from COVID-19 pneumonia. Furthermore, we defined a method for the automatic selection and combination of multi-modal variables towards a holistic signature designed for the COVID-19 triage. On the basis of this interpretable, clinically relevant signature we develop advanced machine learning techniques integrating multi-modal data for severity assessment and short/long term outcome prediction. Our method endows robustness, good generalization properties, explainability and establishes causality with known clinical COVID-19 confounding factors. In conclusion, we show that the combination of chest CT and artificial intelligence can provide tools for fast, accurate and precise disease extent quantification as well as the identification of patients with severe short-term outcomes. Beyond the diagnostic value of CT for COVID-19, our study suggests that AI should be part of the triage process. We are currently working on adapting this pipeline to several other fields such as cancer patients’ response to treatment.
Guillaume Chassagnon, Maria Vakalopoulou, Enzo Battistellaet al., AI-driven quantification, staging and outcome prediction of COVID-19 pneumonia, Medical Image Analysis, Volume 67, 2021
In collaboration with the RICE University under the supervision of Prof. Kavraki.
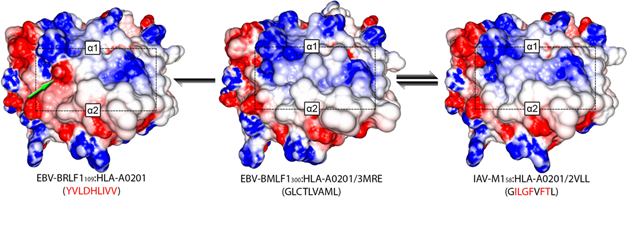
A major challenge in designing immunotherapy is to account for the large variability of possible proteomics profiles of both the particular cancer to treat and of the patient healthy cells. However, most of the methods tackling this task are relying either on the protein sequences that are lacking the ability to consider the spatial structure of the protein or on a biological characterization of the protein structure and then a correlation estimation. We consider HLA proteins for which we have a ground truth dendogram characterizing the similarity between proteins experimentally established. The similarity between the proteins of this dataset has been assessed, in a first step, experimentally by assessing the cross-reactivity to a reference dataset. Graph matching techniques aim to use both information of a graph structure and some features on a graph node to establish a mapping between the nodes of two different graphs. Here, we consider for graphs a mesh on the proteins structures, the nodes being the atoms of the molecule and the edges characterizing spatial proximity of the atoms. The objective of the algorithm is then to maximize the similarity between matched atoms and between the edges of the matched atoms. This objective function is then used to characterize the similarity between the two molecules. Promising results for this proof of concept scenario on the 29 different HLA proteins have been obtained. We achieved an accuracy of 0.83 for the separation of high, low and without cross-reactivity molecules indicating the potentials of our method.
In collaboration with Charlotte Dion.
In order to model the continuous signal of the membrane potential of a neuron by taking into account the signals (spikes) sent by neurons surrounding the neuron of interest, we have proposed a jump diffusion model with jumps driven by a multi-dimensional Hawkes process. For this model we have established ergodicity results (in collaboration with Charlotte Dion and Eva Löcherbach, see [1]), allowing us to make statistical inference on the model parameters. We proposed a drift estimation procedure and established oracle inequalities to guarantee the theoretical performances of our estimator (in collaboration with Charlotte Dion, see [2]). In a second work, we are interested in the estimation of the volatility term and the jump function of the model (in collaboration with Chiara Amorino, Charlotte Dion and Arnaud Gloter). Finally we are studying real data obtained by measuring the membrane potential of a fixed neuron of a turtle as well as the spike trains from a number of neurons around the fixed neuron and looking at how to apply our jump diffusion model to these data (in collaboration with Charlotte Dion and Anna Bonnet).
[1] Dion, C., Lemler, S., & Löcherbach, E. (2019). Exponential ergodicity for diffusions with jumps driven by a Hawkes process. arXiv preprint arXiv:1904.06051, to appear in Theory of Probability and Mathematical Statistics. [2] Dion, C., & Lemler, S. (2019). Nonparametric drift estimation for diffusions with jumps driven by a Hawkes process. Statistical Inference for Stochastic Processes, 1-27

